Administrative Core Project Summary:
This innovative integrated systems biology application seeks to delineate the complex host/pathogen interactions occurring at the alveolar level that lead to unsuccessful response to therapy in serious pneumonia. The Administrative Core will be responsible for managing, coordinating, and supervising the entire range of SCRIPT Systems Biology Center activities. As such, the Administrative Core will intersect with all internal SCRIPT investigators as well as the NIAID sponsoring agency, other Systems Biology Centers, and the broader infectious diseases and pulmonary/critical care scientific communities. The SCRIPT Systems Biology Center is intentionally multidisciplinary, increasing the complexity of interactions and mandating a centralized approach to facilitate accomplishing the goals of this Systems Biology Center.
Data Management and Bioinformatics Core Project Summary:
The overall goal of the Data Management and Bioinformatics (DMBI) Core is to develop and implement new and enhanced computational resources that support a systems biology approach to Hospital Acquired Pneumonia (HAP), and to share those resources broadly. The design and implementation of the DMBI Core is based on optimizing the utility of data generated from both the clinical and laboratory settings – including phenomics, transcriptomics, epigenomics, and meta-genomics. Our systems biology approach affords equal opportunity for discoveries across all data sources to produce significant findings. Achieving the broader goals of this project will require the seamless integration of clinical data with molecular profiling data on both host and pathogen.
Technology Core Project Summary:
The overall goal of the Technology Core is to provide sample processing and data generation support for all projects and other cores in the Successful Clinical Response In Pneumonia Therapy (SCRIPT) Systems Biology Center. Our Technology Core is uniquely poised to contribute to the success of the SCRIPT Study, as we possess expertise in flow cytometry, cell sorting, various Next Generation Sequencing techniques, including RNA-seq for gene expression profiling and genome-wide bisulfite sequencing for genomic DNA methylation, and humanized mouse modeling.
Modeling Core Project Summary:
We will approach the modeling of the development of pneumonia and its resolution in response to treatment using the conceptual framework of bifurcation theory. In the simplest case, with only two states, the model would distinguish a state with low bacterial load and high lung function from one with a high bacterial load and low lung function. The major challenge in this framework is to determine how to express the control parameter in terms of biological variables pertinent to pneumonia pathogenesis. We will pursue an agnostic modeling approach to the challenge of obtaining insight from these high-dimensional data. Because of the complexity of the problem, we will iteratively apply a variety of cutting-edge methods from systems biology, data science, dynamical systems, and ecology. Predictive biomarkers developed in the Modeling Core will then be validated in a prospective confirmatory cohort of patients in whom analogous data will be generated.
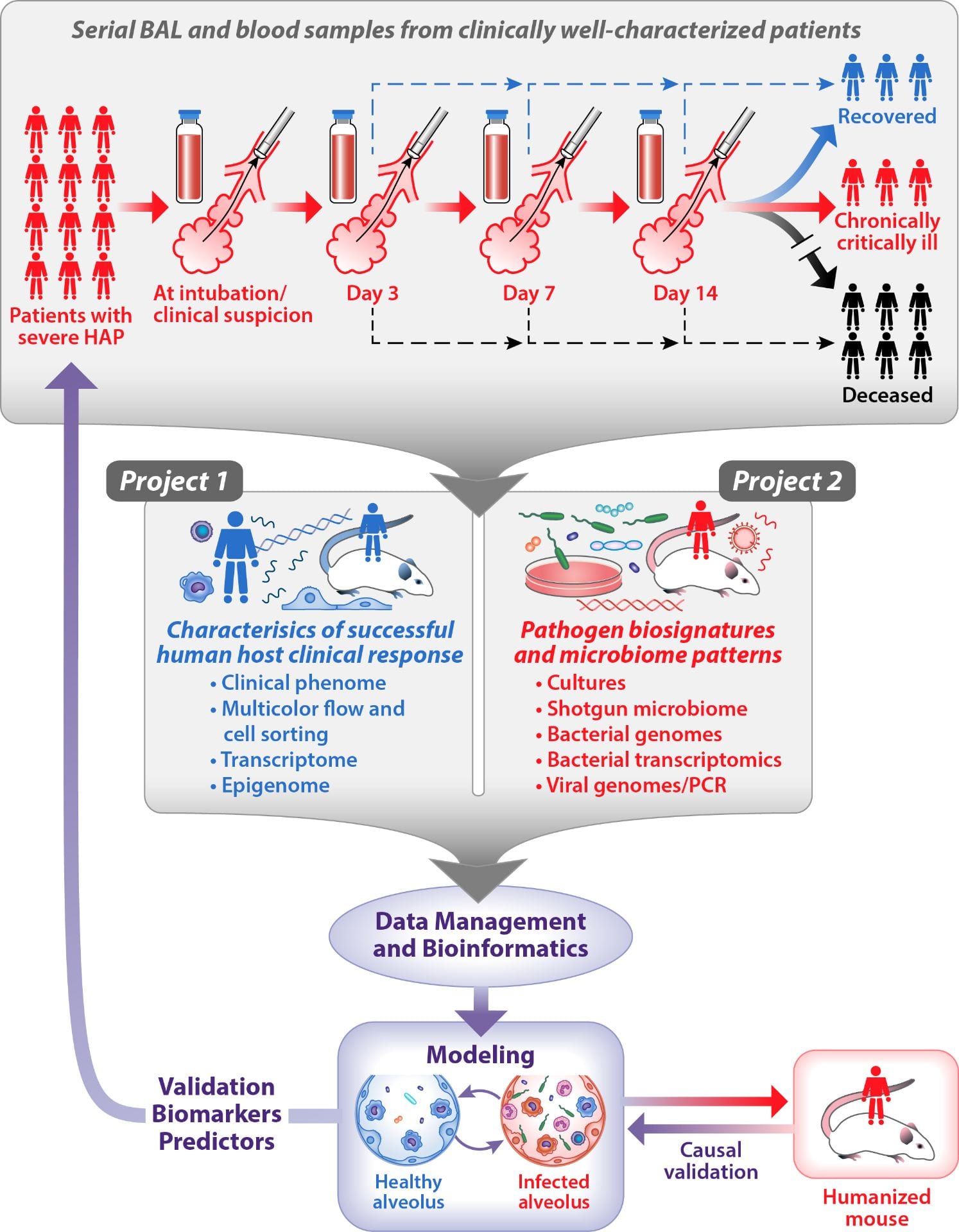
Project 1 Summary: Host response to pneumonia
Hospital-acquired pneumonia (HAP) is the leading causes of death from nosocomial infection, with high rates of associated morbidity. HAP due to Pseudomonas aeruginosa and Acinetobacter spp. is substantially more difficult to treat than other pathogens, with clinical failure rates as high as 50%. These rates persist despite optimization of antibiotic regimens, suggesting factors beyond antibiotic resistance contribute to the persistent morbidity and mortality. In this Project, we will focus on the host response to pneumonia. We hypothesize that persistent inflammation in the alveolus after appropriate antibiotic treatment contributes to clinical failure in patients with P. aeruginosa or Acinetobacter spp. pneumonia.
Project 1 will deploy robust tools for flow sorting macrophage and lymphocyte subset populations, isolating RNA from these populations, and performing transcriptomic and epigenomic analysis to compare successful and unsuccessful host responses to infection.
Project 2 Summary: Microbial determinants of failure of antimicrobial therapy
The overall goal of SCRIPT Research Project 2 is to create a computational model based on microbial biosignatures that predicts clinical failure in patients with hospital-acquired pneumonia. Specific pathogens such as Pseudomonas aeruginosa and Acinetobacter baumannii are particularly problematic in ventilator-associated pneumonia and are associated with clinical failure rates as high as 50%, even in patients treated with appropriate antibiotic therapy. For this reason, a more detailed analysis will be performed on pneumonia caused by these pathogens.
Project 2 will focus on two main areas: 1) the bacterial genomic profiles and 2) the microbiome changes and communities (including bacteria, bacteriophage, other viruses, and fungi) associated with unsuccessful outcomes in patients with ventilator-associated pneumonia.
NBBAL Extraction Process